CREx: examples
here you can find some examples demonstrating the capabilities of CREx. for each example you can download
the gene orders in the fasta format. so you can try it for yourself. first we demonstrate the ability to
localize tdrls and second we demonstrate the usage for a larger data set of echinoderms gene orders.
note that the examples show the original output from crex. we have only omited little details to
simplify the presentation, e.g. the highlighting of the rearrangements in the family diagrams is enabled here
by default, usually you have to trigger the highlighting by clicking on the corresponding rearrangements
in the scenario (but you can try it here also).
the abbreviations of the gene names used in the examples and the example files are documented here
localisation of tdrls
Chauliodus sloani → Eurypharynx pelecanoides
in this example CREx identifies one tdrl (affecting the genes from atp8 to W (in chauliodus sloani)) and
one transposition (of -Y and -C). the tdrl was described in Inoue et al. (2003)
(click on the rearrangements listed in the scenario in order to highlight them). the tdrl is directed
toward the eurypharynx pelecanoides gene order because the other direction needs three tdrls, as shown below.
download fasta .
Chauliodus sloani → Eurypharynx pelecanoides
-
family diagram for Chauliodus sloani
-
family diagram for Eurypharynx pelecanoides
-
scenario:
Eurypharynx pelecanoides → Chauliodus sloani
-
family diagram for Eurypharynx pelecanoides
-
family diagram for Chauliodus sloani
-
scenario:
-
transposition
- prime node scenario
Limulus polyphemus → Narceus annularus, Thyropygus sp
This example was described in detail in Lavrov et al. (2002).
it is an example if "tandem duplication non-random loss", note that all negative genes are moved
to the back by the rearrangement. Again the direction of the tdrl can be inferred with the
parsimony criterion as the other direction needs two tdrl and a transposition.
download fasta
Limulus polyphemus → Narceus annularus, Thyropygus sp
-
family diagram for Limulus polyphemus
-
family diagram for Narceus annularus, Thyropygus sp
-
scenario:
reconstruction of ancestral states
we have reconstructed the ancestral states for the known mitochondrial gene
orders of the echinoderm species (except for the ophiorids) for a
given phylogenetic tree (see Perseke et al. 2007).
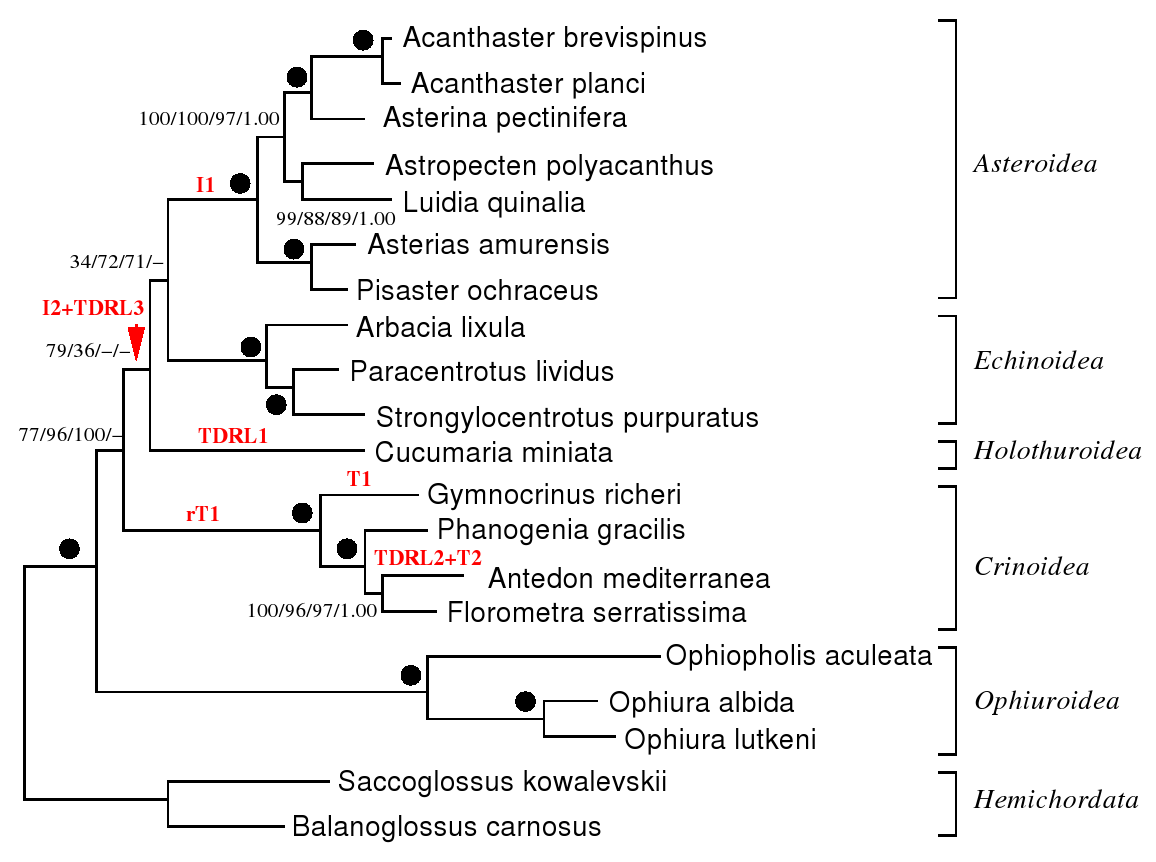
you can find the dataset here. The phylogenetic tree that we have
used is depicted in the following - with the rearrangements annotated to the
edges. the abbreviations are I: inversion, T: transposition, rT: reverse transposition, and TDRL: tandem duplication random loss.
(the dots and numbers can be ignored here - they represent support valued for the
phylogenetic analysis based on molecular data). how the rearrangements were assigned to the edges
is described below.
as you load the data set in CREx you can see immediately that there are a number of equal gene orders
this is:
- all asteroid species (A. brevispinus, P. ochraceus, A. amurensis, L. quinalia, A. polyacanthus, A. pectinifera, A. planci)
- all echinoid species (S. purpuratus, A. lixula, P. lividus)
- the two crinoid species p. gracilis and f. serratissima
- the two ophiuroids o. albidia and o. lutkeni
now lets reconstruct the rearrangement history
- I1: Asteroidea vs. Echinoidea:
the CREx comparison shows the inversion incorporating 17 genes documented by
Asakawa et al. 1995. this gives no information
about the ancestral state of the asteroid-echinoid group, because the inversion
may be located on the asteroid lineage as well as on the echinoid lineage.
to infer the ancestral state we need outgroup information. this is the next step.
Asteroidea → Echinoidea
-
family diagram for Asteroidea
-
family diagram for Echinoidea
-
scenario:
- TDRL1: Echinoidea vs. C. miniata:
the holothuroid can give the information needed to infer the ancestral state of the asteroid-echinoid
group. furthermore - as we will demonstrate - it gives the ancestral state of the
asteroid+echinoid+holothuroid group. the orientation of the genes is the same in echinoids
and the holothuroid and there is one tdrl separating the two groups. therefore
the ancestral state of asteroids+echinoids has to be the echinoid gene order.
because we know the direction of the tdrl (echinoid → holothuroid) we can also infer that
the ancestral state of all three groups is the echinoid gene order.
below you can see the echinoid holothuroid comparison. this is a bit complicated as in C. miniata
there is a duplicated putative control regions annotated (one between the tRNAs E and P and the
other between the tRNAs T and Q). because the current version of CREx can not handle duplicated
genes we show both possibilities here. both show the same TDRL rearrangement
(i.e. both putative control regions are consistent with the rearrangement.)
this example illustrates perfectly - again - the asymetry of TDRL operations
(and the corresponding distance measure): To transform the gene order of
C. miniata (A and B) into the Echinoid order three TDRL rearrangements are needed.
Echinoidea → Cucumaria miniata a
-
family diagram for Echinoidea
-
family diagram for Cucumaria miniata a
-
scenario:
Echinoidea → Cucumaria miniata b
-
family diagram for Echinoidea
-
family diagram for Cucumaria miniata b
-
scenario:
- T1: Florometra serratissima vs. Gymnocrius richeri &
TDRL3 and T2: Florometra serratissima vs. Antedon mediterranea:
Gymnocrius richeri and Florometra serratissima are separated by a transposition. to transform the gene order of
Antedon mediterranea into Florometra serratissima also one transposition (in the same region) and additionally
a tdrl (in the direction of Antedon mediterranea) is needed. with the given tree topology we can
conclude that the gene order of Florometra serratissima is the ancestral order of the crinoids.
Florometra serratissima, Phanogenia gracilis → Gymnocrinus richeri
-
family diagram for Florometra serratissima, Phanogenia gracilis
-
family diagram for Gymnocrinus richeri
-
scenario:
Florometra serratissima, Phanogenia gracilis → Antedon mediterranea
-
family diagram for Florometra serratissima, Phanogenia gracilis
-
family diagram for Antedon mediterranea
-
scenario:
- Echinoidea vs. Florometra serratissima: I2, TDRL3, and rT1
the echinoid gene order is separated from the ancestral crinoid gene order by three rearrangements.
tdrl3 lies on the echinoid lineage (the other direction needs two transpositions). inversion2 and tdrl3
act on the same set of genes. we suppose that those two rearrangements are mechanistically coupled,
which would imply that inversion2 lies also on the echinoid lineage. the reverse transposition
lies on the crinoid lineage as the inversion of the control region is unique to the crinoids
(Scouras et al. 2004).
another - in our opinion less likely scenario - is described in Perseke et al. 2007.
Florometra serratissima, Phanogenia gracilis → Echinoidea
-
family diagram for Florometra serratissima, Phanogenia gracilis
-
family diagram for echinoidea
-
scenario:
-
reverse transposition
- prime node scenario
